The Importance of Traceability


Traceability is crucial in all steps of clinical data handling, from data collection to final analysis. The importance of Traceability in CDISC, and particularly in ADaM, is continuously “stressed upon” throughout the CDISC documentation (CDISC “ADaM Traceability Examples”; Document will be released in Q4-2021) and at conferences[1,2,3,4,5] , including a recent FDA presentation at CDISC EU 2021 European Interchange[6] .
For those of you who are not yet familiar with the concept of traceability, I will use the CDISC definition and an excerpt from the FDA Study Data Technical Conformance Guidance:
CDISC: “Traceability is the property that enables the understanding of the data’s lineage and/or the relationship between an element and its predecessor(s). Traceability is built by clearly establishing the path between an element and its immediate predecessor”
FDA: “Establishing traceability is one of the most problematic issues associated with any data conversion. If the reviewer is unable to trace study data from the data collection of subjects participating in a study to the analysis of the overall study data, then the regulatory review of a submission may be compromised” … the last part of this sentence looks like if not yet a threat, it is at least a warning.
Traceability in ADaM can be achieved in two ways, through data-point and through metadata.
Data-Point Traceability
Retaining the --SEQ variable from SDTM source dataset is a starting point. Below is a simple example on how to achieve data-point traceability between an ADaM dataset (ADAE) and its predecessor (SDTM AE dataset), i.e., by keeping in the ADaM dataset the sequence variable from the SDTM dataset (AESEQ).
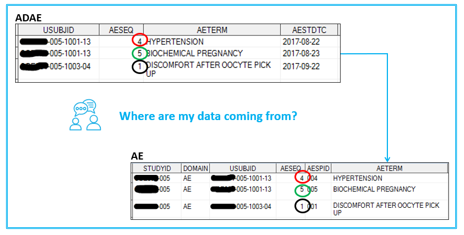
Of course, there is more to it than that and the ADaM IG provides examples on what other methods you can use to maintain data traceability through your ADaM datasets. If an ADaM dataset is well designed using the appropriate ADaM rules and best practices, then by just opening an ADaM dataset a reviewer should, to a certain extent, understand what kind of alterations have been made to which data. This can be achieved through other more sophisticated data-point traceability techniques. For example:
- When you need to re-derive in ADaM, a variable collected in SDTM (e.g., AGE), create a new variable and keep original variables in the ADaM dataset
- When you need to derive a variable using variable(s) from SDTM, keep original SDTM variable(s) used in the derivation
- When you need to impute a missing or partial value e.g., date, create a new variable and keep the original SDTM variable. For date/time variable makes use of imputation flag variables (e.g., ADTF)
- When applying windowing e.g., create AVISIT (Analysis Visit) keep “supporting” ADaM windowing variables and keep original SDTM timepoint variable(s) (e.g., VISITNUM)
- When you need to impute a missing timepoint from other timepoints e.g., visit, derive a new record (use DTYPE) and keep original timepoints records even if not analyzed. Make use of flags (e.g., ANLxxFL) to identify records/visits used in the analysis vs those not used
- When you deal with complex derivations requiring several steps, make use of Intermediate datasets[5]
Metadata Traceability
Together with data traceability, or whenever data traceability is complex to implement, we can use metadata traceability. This essentially means, to describe the define-xml through your metadata, and also in the supporting documentation, e.g., the Analysis Data Reviewer Guide, mention the transformations that your data went through:
- An algorithm used for deriving a variable
- An entirely “new” derived analysis concept, e.g., progression free survival in Oncology
- A visit windowing, e.g., when data are coming from external labs and the study visit is not available but only the sample/assessment date imputations, either for a single variable or for an entire time-point, e.g., when a subject discontinued prematurely
- identification of records not used in the analysis, e.g., all efficacy assessments measured after the use of a rescue or a prohibited medication
The CDISC ADaM Team should release in Q4-2021, a document where additional examples of traceability will be provided (the document has already gone through the public review process).
The document that was out for review earlier this year, contains 12 instances of fully explained metadata samples together with outputs and ADaM datasets examples:
- General ADSL Traceability
- Traceability with Parameters from Multiple Input Datasets
- Traceability When Multiple Datasets Are Merged
- Traceability When Multiple Input Datasets are Stacked to Create OCCDS
- Traceability When Using a Look-Up Table
- Traceability When Adding a Row to a BDS Dataset
- Traceability When Multiple Analysis Variables are needed on the Same Row
- Using an Intermediate Dataset for BDS Traceability / Using an Intermediate Dataset for ADSL Traceability
If you want to know more on how to achieve traceability in ADaM, join me at the next PHUSE EU Connect, 15th - 19th November 2021 (Click here for Agenda), where I will present during the Standard Implementation stream on the topic “How to be Traceable in ADaM”. Click here to register for the virtual event.
References:
[1] “Traceability: Some Thoughts and Examples for ADaM Needs", S. Minjoe, Q. Zhou and R. Watson; PharmaSUG, 2018
[2] “Lost in Traceability- From SDTM to ADaM” Cytel Blog https://www.cytel.com/blog/lost-in-traceability, 2016
[3] “« Lost » in Traceability, from SDTM to ADaM …. finally, Analysis Results Metadata”, A. Tinazzi; CDISC EU Interchange, 2016
[4] “ADaM Intermediate Dataset: how to improve your analysis traceability”, A. Tinazzi, T. Curto and A. Aggarwal; PHUSE US Connect, 2018
[5] “Leveraging Intermediate Data Sets to Achieve ADaM Traceability”, Y. Zhuo; PharmaSUG, 2019
[6] “CDER’S Experience with the ADaM Traceability Assessment and Common Data Quality Issues”, J. Anderson; CDISC EU Interchange, 2021
About Angelo Tinazzi
 Angelo Tinazzi is Senior Director, Statistical Programming, Clinical Data Standards and Clinical Data Submission at Cytel. He is a well- published and recognized expert in statistical programming with over 20 years' experience in clinical research. The application of CDISC standards in different therapeutic areas is part of his core expertise since 2003 in particular in the context of data submission to health authorities such as the FDA and PMDA.
Angelo Tinazzi is Senior Director, Statistical Programming, Clinical Data Standards and Clinical Data Submission at Cytel. He is a well- published and recognized expert in statistical programming with over 20 years' experience in clinical research. The application of CDISC standards in different therapeutic areas is part of his core expertise since 2003 in particular in the context of data submission to health authorities such as the FDA and PMDA.
Angelo is an authorized CDISC instructor and member of the CDISC ADaM Team as well as the CDISC European Committee where he also manages the Italian-speaking CDISC User Network.

